Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
hanging drop vapor diffusion method, using 1 M sodium potassium tartrate, 200 mM sodium chloride, 100 mM imidazole pH 8.0
hanging drop vapor diffusion method, using 0.25 M NaCl, 28% (w/v) polyethylene glycol 3350 and 0.1 M bis-Tris, pH 5.5
hanging-drop vapor-diffusion method at 20°C, crystal structure in complex with NADP+ and two Mg2+ ions at 2.6 A resolution
in complex with Mg2+ and NADPH at 2.3 A resolution. The binding of Mg2+ increases structural disorder while the binding of NADPH increases the structural rigidity of the enzyme. The binding of Mg2+ and NADPH opens the interface between the N- and C-domains, thereby allowing access for the substrates to bind
purified recombinant His-tagged enzyme, 9 mg/ml protein in 20 mM sodium HEPES, pH 7.5, and NADPH in a ratio of 10 mol NADPH per mol of enzyme, hanging drop vapour diffusion method, equal volumes of 0.003 ml of protein and reservoir solution, the latter containing 1.6 M ammonium sulfate, and 0.1 M sodium bicine, pH 9.0, 17°C, 6 months, X-ray diffraction structure determination and analysis at 2.6 A resolution
-
the enzyme is remarkably easy to crystallize
-
hanging drop vapor diffusion method, using 0.1 M bis-Tris, pH 6.0, 22% (w/v) polyethylene glycol monomethylether 5000
in complex with NADPH and the transition state analogue N-isopropyloxamate and apo-form. The enzyme has a seven-residue specificity loop
native enzyme with two magnesium ions bound in the active site, hanging drop vapor diffusion method, using 250 mM MgCl2, 20% (w/v) PEG 3350, 100 mM Tris/HCl, pH 8.0
enzyme KARI in complex with Mg2+ or with Mg2+ and NADPH, hanging-drop method by vaporphase diffusion at 18°C, 0.003 ml of protein solution containing 6 mg/ml enzyme, 50 mM Hepes, pH 7.5, 5 mM NADPH, and 5 mM MgCl2, is mixed with 0.001 ml of reservoir solution containing 0.2 M magnesium chloride hexahydrate, 0.1 M Tris-HCl, pH 8.5, and 15% w/v PEG 4000, a few days to 3 months, X-ray diffraction structure determination and analysis at 1.55 A and 2.80 A resolution, respectively, molecular replacement
The dodecamer architecture of 23 point group symmetry is assembly of six dimeric units and dimerization is essential for the formation of the active site
-
X-ray, structure analysis
-
ammonium sulfate precipitation
-
ammonium sulfate precipitation, crystal structure of the enzyme complexed with NADPH, two magnesium ions and N-hydroxy-N-isopropyloxamate, a herbicidal transition state analog determined at 1.65 A resolution, recombinant enzyme overexpressed in Escherichia coli
-
crystallized with 2-aceto-2-hydroxybutanoate, Mn2+ and NADPH
-
crystal structures of the enzyme in complex with two transition state analogues, cyclopropane-1,1-dicarboxylate and N-isopropyloxalyl hydroxamate. Hanging-drop vapor-diffusion method
hanging drop method, cocrystallization of 3-((methylsulfonyl)methyl)-2H-benzo[b][1,4]oxazin-2-one in complex with the enzyme
hanging-drop vapor-diffusion method. Crystal structures of Staphylococcus aureus KARI in complex with two transition state analogues, cyclopropane-1,1-dicarboxylate and N-isopropyloxalyl hydroxamate
micro-batch and hanging-drop vapor-diffusion methods. The crystal structure is determined at 1.69 A resolution. The crystal structure of SpIlvC contains an asymmetric dimer in which one subunit is in apo-form and the other in NADP(H) and Mg2+-bound form
crystal structure at a 1.75 A resolution
hanging drop vapor diffusion method, using 0.1 M bis-Tris, pH 5.0, 20% (w/v) w/v polyethylene glycol 1500
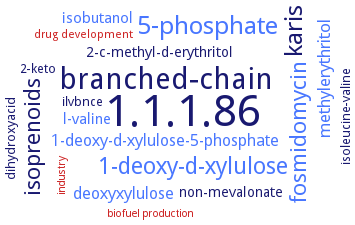



 results (
results ( results (
results ( top
top





