3.2.2.19: [protein ADP-ribosylarginine] hydrolase
This is an abbreviated version!
For detailed information about [protein ADP-ribosylarginine] hydrolase, go to the full flat file.
Word Map on EC 3.2.2.19 
Reaction
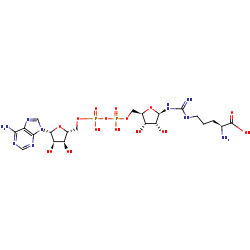 Nomega-(ADP-D-ribosyl)-L-arginine +
Nomega-(ADP-D-ribosyl)-L-arginine +
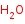 H2O=
H2O=
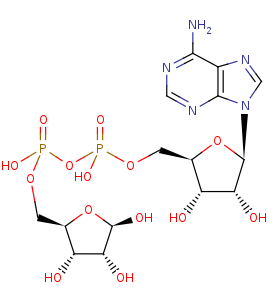 ADP-D-ribose +
ADP-D-ribose +
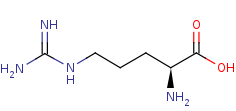 L-arginine
L-arginine
Synonyms
AAH, ADP-ribose-L-arginine cleavage enzyme, ADP-ribose-L-arginine cleaving enzyme, ADP-ribosyl(arginine)protein hydrolase, ADP-ribosyl-acceptor hydrolase, ADP-ribosylarginine glycohydrolase, ADP-ribosylarginine hydrolase, ADP-ribosylhydrolase, ADP-ribosylhydrolase 1, ADP-ribosylhydrolase 3, ADP-ribosylhydrolase-like 1, ADPRH, AdprHl1, ARH1, ARH3, hARH1, mono-ADP-ribosyl-arginine hydrolase, More, Nomega-(ADP-D-ribosyl)-L-arginine ADP-ribosylhydrolase, O-acetyl-ADP-ribose hydrolase, PARG, poly(ADP-ribose) glycohydrolase, poly-(ADP-ribose) glycohydrolase, [protein ADP-ribosylarginine] hydrolase
ECTree
Sequence
Sequence on EC 3.2.2.19 - [protein ADP-ribosylarginine] hydrolase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
ADPRH_BOVIN
353
0
39152
Swiss-Prot
Secretory Pathway (Reliability: 5)
ADPRH_DICDI
392
0
44362
Swiss-Prot
other Location (Reliability: 2)
ADPRH_HUMAN
357
0
39507
Swiss-Prot
other Location (Reliability: 3)
ADPRH_MOUSE
362
0
40068
Swiss-Prot
Secretory Pathway (Reliability: 5)
ADPRH_RAT
362
0
39961
Swiss-Prot
Secretory Pathway (Reliability: 5)
TRI1_PSEP8
Pseudomonas putida (strain DSM 28064 / B6-2)
382
0
41685
Swiss-Prot
-
TRI1_PSEPG
Pseudomonas putida (strain GB-1)
371
0
40782
Swiss-Prot
-
TRI1_SERP5
Serratia proteamaculans (strain 568)
366
0
40934
Swiss-Prot
-
C0RGH1_BRUMB
Brucella melitensis biotype 2 (strain ATCC 23457)
359
0
39601
TrEMBL
-
T0YYQ3_9ZZZZ
279
0
30422
TrEMBL
other Location (Reliability: 2)
A0A6G9Q4D9_LENHI
319
0
35850
TrEMBL
-
A0A7S4H458_9GAMM
363
0
40251
TrEMBL
-
A0A2N9NXA1_9BACT
313
0
33238
TrEMBL
-
A0A0N0D583_9DELT
261
0
29370
TrEMBL
-
M5TC16_9BACT
477
0
54023
TrEMBL
-
Q66IX9_XENLA
353
0
39057
TrEMBL
Secretory Pathway (Reliability: 5)
A0A6V6YX14_9FLAO
Flavobacterium salmonis
312
0
35187
TrEMBL
-
C0G3N9_9HYPH
404
0
44435
TrEMBL
-
A0A6J5E934_9BURK
308
0
32546
TrEMBL
-
T1CST5_9ZZZZ
344
0
37090
TrEMBL
other Location (Reliability: 2)
E3TEX4_ICTPU
352
0
38370
TrEMBL
other Location (Reliability: 2)
A0A176S542_9GAMM
32
0
3406
TrEMBL
-
A0A3M7SI05_BRAPC
352
0
39787
TrEMBL
other Location (Reliability: 2)
A0A1X7MB84_9GAMM
methanotrophic bacterial endosymbiont of Bathymodiolus sp
135
0
15103
TrEMBL
-
K1RMX6_9ZZZZ
115
0
12528
TrEMBL
other Location (Reliability: 1)
A0A6J8E5T5_MYTCO
459
0
50513
TrEMBL
Secretory Pathway (Reliability: 4)
E4PJ11_MARAH
Marinobacter adhaerens (strain DSM 23420 / HP15)
390
0
41837
TrEMBL
-
A0A0F0CQF5_9BACT
3394
0
389185
TrEMBL
-
J9FP61_9ZZZZ
252
0
28318
TrEMBL
other Location (Reliability: 2)
K1SMY5_9ZZZZ
396
0
44612
TrEMBL
Secretory Pathway (Reliability: 5)
F2AZH5_RHOBT
297
0
32026
TrEMBL
-
K5DCF1_RHOBT
345
0
37386
TrEMBL
-
A0A176RSG0_9GAMM
133
0
14642
TrEMBL
-
A9ULD9_XENTR
353
0
39106
TrEMBL
other Location (Reliability: 4)
M5T5U6_9BACT
352
0
38564
TrEMBL
-
A0A822II52_9EURY
339
0
37058
TrEMBL
-
K1TRD3_9ZZZZ
224
0
25441
TrEMBL
other Location (Reliability: 3)
A0A6J8E1S3_MYTCO
459
0
50704
TrEMBL
other Location (Reliability: 5)
T1AZ80_9ZZZZ
360
0
38535
TrEMBL
other Location (Reliability: 2)
A0A1F2P6K1_9EURY
309
0
32734
TrEMBL
-
T1BSY4_9ZZZZ
148
0
15033
TrEMBL
other Location (Reliability: 4)
A0A6V6Y078_9FIRM
256
0
28337
TrEMBL
-
M5U4F7_9BACT
350
0
37505
TrEMBL
-
M2B4B0_9BACT
354
0
38098
TrEMBL
-
A0A176RVV5_9GAMM
255
0
28635
TrEMBL
-
A0A176S1L0_9GAMM
282
0
31288
TrEMBL
-
A0A6S7BRU3_9BURK
324
0
35573
TrEMBL
-
C4WGS7_9HYPH
367
0
40461
TrEMBL
-
K1UKE2_9ZZZZ
199
0
22698
TrEMBL
other Location (Reliability: 2)
L7CBM4_RHOBT
345
1
37531
TrEMBL
-
G8PVM6_PSEUV
Pseudovibrio sp. (strain FO-BEG1)
319
0
34747
TrEMBL
-
K1T0M8_9ZZZZ
265
0
29905
TrEMBL
other Location (Reliability: 2)
K5CC60_RHOBT
297
0
31941
TrEMBL
-
A0A6J8E4C7_MYTCO
218
0
24152
TrEMBL
other Location (Reliability: 2)
A0A822INH9_9EURY
319
0
34777
TrEMBL
-
A0A1F2P9D2_9EURY
308
0
32758
TrEMBL
-
T1BTS0_9ZZZZ
360
0
38516
TrEMBL
other Location (Reliability: 2)
Q5U799_AZOLI
312
0
33898
TrEMBL
-
A0A8B6D6M8_MYTGA
464
0
51309
TrEMBL
other Location (Reliability: 5)
T1AL83_9ZZZZ
360
0
38543
TrEMBL
other Location (Reliability: 3)
A0A7J5WVH8_9ACTN
357
0
37457
TrEMBL
-
C4IP10_BRUAO
404
0
44435
TrEMBL
-
Q6DBR3_DANRE
353
0
38772
TrEMBL
other Location (Reliability: 3)
A0A822JG07_9EURY
329
0
35601
TrEMBL
-
F2B1F2_RHOBT
354
0
38431
TrEMBL
-
M5RTY7_9BACT
355
0
38249
TrEMBL
-
A0A6V6YQW6_9FLAO
315
0
35589
TrEMBL
-
YXEE_BACSU
Bacillus subtilis (strain 168)
121
0
14714
Swiss-Prot
-
ARHL1_XENLA
354
0
40205
Swiss-Prot
-
html completed




 results (
results ( results (
results ( top
top









