6.3.2.9: UDP-N-acetylmuramoyl-L-alanine-D-glutamate ligase
This is an abbreviated version!
For detailed information about UDP-N-acetylmuramoyl-L-alanine-D-glutamate ligase, go to the full flat file.
Reaction
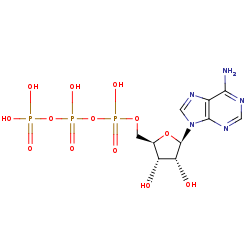 ATP +
ATP +
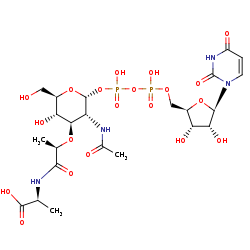 UDP-N-acetyl-alpha-D-muramoyl-L-alanine +
UDP-N-acetyl-alpha-D-muramoyl-L-alanine +
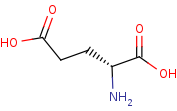 D-glutamate=
D-glutamate=
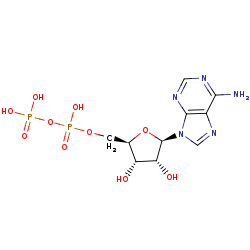 ADP +
ADP +
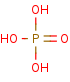 phosphate +
phosphate +
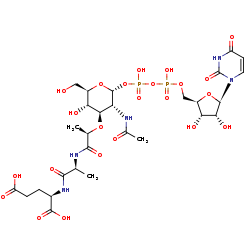 UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-D-glutamate
UDP-N-acetyl-alpha-D-muramoyl-L-alanyl-D-glutamate
Synonyms
D-Glutamate-adding enzyme, D-glutamic acid adding enzyme, D-Glutamic acid-adding enzyme, MurD, MurD cell wall enzyme, MurD ligase, Synthetase, uridine diphospho-N-acetylmuramoylalanyl-D-glutamate, UDP-Mur-Nac-L-Ala:L-Glu ligase, UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase, UDP-N-acetylmuramoyl-L-alanine:D-glutamateligase, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate synthetase, UDPMurNAc-L-alanine:D-glutamate ligase (ADP-forming), Uridine diphosphate N-acetylmuramoyl-L-alanine:D-glutamate ligase, Uridine diphospho-N-acetylmuramoylalanyl-D-glutamate synthetase
ECTree
Metals Ions
Metals Ions on EC 6.3.2.9 - UDP-N-acetylmuramoyl-L-alanine-D-glutamate ligase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
arsenate
-
increases storage stability
Ca2+
-
increases storage stability
Cd2+
-
abolishes the enzyme activity at 5mM
phosphate
-
increases storage stability
potassium phosphate
-
highly dependent on, optimal concentration: 11-16 mM, inhibition above 20 mM
sulfate
-
increases storage stability
vanadate
-
abolishes the enzyme activity at 5mM
Zn2+
-
abolishes the enzyme activity at 5mM
Mg2+

-
optimal concentration: 5-25 mM for the spore enzyme, 6-28 mM for the enzyme from vegetative cells
Mg2+
-
highly dependent on, optimal concentration: 5 mM
Mg2+
-
increases storage stability
Mg2+
-
optimal concentration: 10 mM
Mg2+
-
optimal concentration: 10 mM, independent of ATP concentration
Mg2+
-
required, optimal concentration 15 mM
Mn2+

-
increases storage stability
Mn2+
-
absolute requirement, maximal activity at concentrations of Mn2+ equimolar to ATP
Mn2+
-
optimal concentration: 1 mM



 results (
results ( results (
results ( top
top






