4.4.1.21: S-ribosylhomocysteine lyase
This is an abbreviated version!
For detailed information about S-ribosylhomocysteine lyase, go to the full flat file.
Reaction
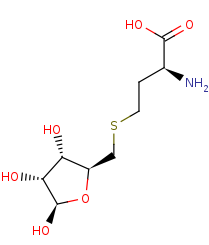 S-(5-deoxy-D-ribos-5-yl)-L-homocysteine=
S-(5-deoxy-D-ribos-5-yl)-L-homocysteine=
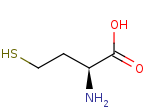 L-homocysteine +
L-homocysteine +
 (4S)-4,5-dihydroxypentan-2,3-dione
(4S)-4,5-dihydroxypentan-2,3-dione
Synonyms
AI-2 synthase, autoinducer-2 synthase, BsLuxS, EC 3.13.1.2, EC 3.2.1.148, EC 3.3.1.3, EcLuxS, lsrR, Lux S, LuxS, LuxS protein, S-ribosyl homocysteinase, S-ribosylhomocysteinase, S-ribosylhomocysteine lyase, S-ribosylhomocysteinelyase, VhLuxS
ECTree
Organism
Organism on EC 4.4.1.21 - S-ribosylhomocysteine lyase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
-
-
-
brenda
-
-
-
brenda
isolate SSU
Uniprot
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
BSIP1758 mutant lacking luxS
SwissProt
brenda
gene luxS
-
-
brenda
gene luxS
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
gene luxS
UniProt
brenda
-
-
-
brenda
no activity in Agrobacterium tumefaciens
-
-
-
brenda
no activity in Alphaproteobacteria
-
-
-
brenda
no activity in Bacteroidetes
-
-
-
brenda
no activity in Desulfovibrio desulfuricans
-
-
-
brenda
no activity in Enterobacter aerogenes
-
-
-
brenda
no activity in Escherichia coli
-
-
-
brenda
no activity in Klebsiella pneumoniae
-
-
-
brenda
no activity in Marinomonas sp.
-
-
-
brenda
no activity in Marinomonas sp. MED121
-
-
-
brenda
no activity in Neptuniibacter caesariensis
-
-
-
brenda
no activity in Pseudomonas aeruginosa
-
-
-
brenda
no activity in Rhodobacter capsulatus
-
-
-
brenda
no activity in Rhodobacter sphaeroides strain 2.4.1
-
-
-
brenda
no activity in Salmonella typhimurium
-
-
-
brenda
no activity in Sinorhizobium meliloti
-
-
-
brenda
no activity in Vibrio vulnificus
-
-
-
brenda
no activity in Zymomonas mobilis
-
-
-
brenda
gene luxS
UniProt
brenda
gene luxS
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
Serratia kiliensis
-
-
-
brenda
-
-
-
brenda
Serratia malilotii
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
Serratia putrefaciens
-
-
-
brenda
strain DSM 9167T
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
serotype 2, gene luxS
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
BSIP1758 mutant lacking luxS
SwissProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
EHEC, O157:H7
SwissProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
isoform LuxS1
ETF12584
GenBank
brenda
isoform LuxS2
ETF12641
GenBank
brenda
isoform LuxS1
ETF12584
GenBank
brenda
isoform LuxS2
ETF12641
GenBank
brenda
-
UniProt
brenda
gene luxS
UniProt
brenda
strains ATCC 55730, LTH6560, and 100-23C
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
gene luxS
-
-
brenda
-
UniProt
brenda
serotypes M1 and M19
-
-
brenda
-
UniProt
brenda
serotype 2, gene luxS
UniProt
brenda
serotype 2, strain HA9801
-
-
brenda
-
-
-
brenda
strain ATCC 33787 and isolate MVP01
UniProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
strains BB170 ATCC BAA1117 and BB152 ATCC BAA1119
-
-
brenda




 results (
results ( results (
results ( top
top







