4.4.1.2: homocysteine desulfhydrase
This is an abbreviated version!
For detailed information about homocysteine desulfhydrase, go to the full flat file.
Reaction
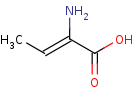 2-aminobut-2-enoate=
2-aminobut-2-enoate=
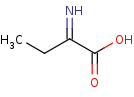 2-iminobutanoate
2-iminobutanoate
Synonyms
desulfhydrase, homocysteine, homocysteine desulfurase, Hyc gamma-lyase
ECTree
Sequence
Sequence on EC 4.4.1.2 - homocysteine desulfhydrase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
MEGL_FUSNN
Fusobacterium nucleatum subsp. nucleatum (strain ATCC 25586 / DSM 15643 / BCRC 10681 / CIP 101130 / JCM 8532 / KCTC 2640 / LMG 13131 / VPI 4355)
395
0
43306
Swiss-Prot
-
MEGL_FUSNP
395
0
43288
Swiss-Prot
-
MEGL_PORGI
Porphyromonas gingivalis (strain ATCC BAA-308 / W83)
399
0
43262
Swiss-Prot
-
MEGL_PSEPU
398
0
42627
Swiss-Prot
-
MEGL_STRAW
Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680)
413
0
43027
Swiss-Prot
-
MEGL_TREDE
Treponema denticola (strain ATCC 35405 / DSM 14222 / CIP 103919 / JCM 8153 / KCTC 15104)
401
0
43488
Swiss-Prot
-
CGL_DICDI
387
0
42497
Swiss-Prot
other Location (Reliability: 2)
CGL_HUMAN
405
0
44508
Swiss-Prot
other Location (Reliability: 2)
CGL_MACFA
405
0
44521
Swiss-Prot
other Location (Reliability: 2)
CGL_MOUSE
398
0
43567
Swiss-Prot
other Location (Reliability: 3)
CGL_PIG
405
0
44490
Swiss-Prot
other Location (Reliability: 2)
CGL_RAT
398
0
43605
Swiss-Prot
other Location (Reliability: 3)
MCCB_BACSU
Bacillus subtilis (strain 168)
379
0
40886
Swiss-Prot
-
A0A0G3WDB8_9CLOT
385
0
42937
TrEMBL
-
A0A0H4NXL3_9BACI
389
0
42927
TrEMBL
-
A0A653MMN0_9MICC
387
0
41269
TrEMBL
-
A0A7R8NVR0_9ACTN
381
0
40082
TrEMBL
-
A0A653T3J2_9MICC
395
0
41401
TrEMBL
-
A0A1V4SF60_RUMHU
394
0
43457
TrEMBL
-
A0A7G2D1R0_9MICO
386
0
40658
TrEMBL
-
A0A653INV2_9MICC
419
0
43551
TrEMBL
-
A0A653TUW6_9BACI
382
0
41430
TrEMBL
-
A0A2U3PK91_LEUCA
380
0
41395
TrEMBL
-
A0A1W7LLI9_CLOBE
386
0
42464
TrEMBL
-
A0A087CFU2_9BIFI
408
0
44185
TrEMBL
-
A0A8H8YBR1_LACPA
379
0
40839
TrEMBL
-
A0A0U1MFC1_STAAU
380
0
41264
TrEMBL
-
A0A2R8A4D4_CARDV
381
0
41063
TrEMBL
-
A0A7W8IQQ9_9BACI
377
0
41159
TrEMBL
-
A0A087A8U8_9BIFI
406
0
44155
TrEMBL
-
J1H1P8_9CLOT
379
0
41896
TrEMBL
-
K4ZQA3_PAEA2
Paenibacillus alvei (strain ATCC 6344 / DSM 29 / NBRC 3343 / NCIMB 9371 / NCTC 6352)
395
0
44080
TrEMBL
-
A0A1S8N4M8_CLOSA
382
0
42376
TrEMBL
-
A0A7W0BTS4_9BACI
377
0
41180
TrEMBL
-
A0A0D8BP60_GEOKU
377
0
41176
TrEMBL
-
A0A087CYW3_9BIFI
415
0
45141
TrEMBL
-
A0A087DP53_9BIFI
406
0
44184
TrEMBL
-
A0A1S1VA22_9FIRM
379
0
41615
TrEMBL
-
A0A840DW42_9BACI
377
0
41212
TrEMBL
-
I4EC89_9CHLR
412
0
44802
TrEMBL
-
A0A852TCK3_9BACI
377
0
40740
TrEMBL
-
A0A8I1XUG2_9BACL
376
0
40949
TrEMBL
-
A0A1F2PFU2_9FIRM
381
0
42221
TrEMBL
-
A0A7W8IIN4_9BACT
386
0
41332
TrEMBL
-
A0A1J5SIB3_9ZZZZ
386
0
41267
TrEMBL
Mitochondrion (Reliability: 3)
A0A6L5B6B8_9BACI
375
0
40789
TrEMBL
-
A0A259V1L1_9FIRM
396
0
43828
TrEMBL
-
A0A841PU51_9BACI
377
0
41256
TrEMBL
-
A0A654APG2_9MICC
387
0
40472
TrEMBL
-
A0A0X8VD99_ANAPI
392
0
43206
TrEMBL
-
A0A653T4Z7_9MICC
387
0
40679
TrEMBL
-
A0A087EBQ0_9BIFI
406
0
44003
TrEMBL
-
A0A1J5P5X8_MOOTH
378
0
40721
TrEMBL
-
A0A090ZIR5_9BACI
377
0
41224
TrEMBL
-
A0A1S8LPD7_9CLOT
381
0
42269
TrEMBL
-
M5RAS1_BACIT
388
0
42218
TrEMBL
-
A0A1D7ZZQ0_LIMFE
381
0
41177
TrEMBL
-
M1MR76_9CLOT
385
0
42528
TrEMBL
-
A0A199XSC7_9FLAO
402
0
43739
TrEMBL
-
I0R6N5_9FIRM
379
0
41693
TrEMBL
-
A0A1S8LGQ8_9CLOT
385
0
43187
TrEMBL
-
A0A0N8NTZ1_9CLOT
383
0
42967
TrEMBL
-
A0A259UH23_9FIRM
380
0
41059
TrEMBL
-
A0A1V4IRP2_9CLOT
381
0
41984
TrEMBL
-
A0A2N9NCT4_9BACT
385
0
41964
TrEMBL
-
A0A1W6VPG0_GEOTD
392
0
43202
TrEMBL
-
A0A1J5NFL4_MOOTH
378
0
40933
TrEMBL
-
A0A259UGI8_9FIRM
381
0
41282
TrEMBL
-
A0A0B4WBP3_CLOBO
380
0
41999
TrEMBL
-
A0A7W8J5K6_9BACT
386
0
41379
TrEMBL
-
A0A811IFA9_STRTR
394
0
43126
TrEMBL
-
U5ML51_CLOSA
382
0
42365
TrEMBL
-
A0A7W0BXH2_9BACI
377
0
41143
TrEMBL
-
A0A1W7LHM8_CLOBE
382
0
42453
TrEMBL
-
A0A7W5C8Q8_9BACL
381
0
41474
TrEMBL
-
A0A166EKX2_9EURY
398
0
44223
TrEMBL
-
A0A087BK54_9BIFI
404
0
44460
TrEMBL
-
A0A7W8N5M7_9BACI
377
0
41154
TrEMBL
-
I0JJR5_HALH3
Halobacillus halophilus (strain ATCC 35676 / DSM 2266 / JCM 20832 / KCTC 3685 / LMG 17431 / NBRC 102448 / NCIMB 2269)
378
0
41366
TrEMBL
-
A0A7W7ZQL3_9BACT
382
0
41218
TrEMBL
-
D5DMS2_PRIM3
Priestia megaterium (strain DSM 319 / IMG 1521)
377
1
41215
TrEMBL
-
A0A653IX69_MICLU
419
0
43504
TrEMBL
-
A0A1V4SM78_RUMHU
388
0
42842
TrEMBL
-
A0A7W0C4C5_9BACI
377
0
41191
TrEMBL
-
L7ZZJ4_9BACI
377
0
41046
TrEMBL
-
A0A1S8RM13_CLOBE
382
0
42323
TrEMBL
-
A0A1V4J1Q6_9CLOT
382
0
42398
TrEMBL
-
A0A212LUJ9_9FIRM
uncultured Sporomusa sp
381
0
41287
TrEMBL
-
A0A6S6XEV0_9FIRM
379
0
42275
TrEMBL
-
A0A7U7JSM5_9STAP
380
0
41313
TrEMBL
-
A0A7X0HSE0_9BACI
378
0
41069
TrEMBL
-
A0A6F9E2D8_9BACL
393
0
42812
TrEMBL
-
A0A1S8SFI2_CLOBE
382
0
42420
TrEMBL
-
A0A1S8KXE0_9CLOT
385
0
43186
TrEMBL
-
R4YVR0_9ACTN
406
0
42257
TrEMBL
-
A0A653VHJ2_9ACTN
396
0
41715
TrEMBL
-
A0A653NMK3_9MICO
384
0
40187
TrEMBL
-
A0A087B6G6_9BIFI
403
0
44267
TrEMBL
-
A0A7X0DAV9_9BACI
377
0
41062
TrEMBL
-
A0A0C1R733_9CLOT
380
0
42144
TrEMBL
-
A0A1U7MJ55_9FIRM
381
0
41287
TrEMBL
-
A0A084GGN6_PSEDA
444
0
47960
TrEMBL
other Location (Reliability: 2)
A0A653Q4U1_9ACTN
391
0
41174
TrEMBL
-
A0A1C4G0R2_BACTU
377
0
41260
TrEMBL
-
A0A654CN90_9STAP
381
0
41641
TrEMBL
-
A0A653M654_9MICO
392
0
41182
TrEMBL
-
A0A2I2KKD7_9ACTN
397
0
41101
TrEMBL
-
A0A841RJM7_9BACI
377
0
41292
TrEMBL
-
A0A2U3KU28_9BACT
385
0
41351
TrEMBL
-
A0A7W8CQ24_9BACL
Planococcus koreensis
377
0
40301
TrEMBL
-
E3HFJ8_ACHXA
Achromobacter xylosoxidans (strain A8)
414
0
44357
TrEMBL
-
F7S8N9_9PROT
239
0
25009
TrEMBL
-
A0A1G4EVY9_BACMY
377
0
41190
TrEMBL
-
A0A076P2V0_FLAPS
400
0
43396
TrEMBL
-
A0A0K2YDK3_9NOCA
384
0
40479
TrEMBL
-
A0A1S8L677_9CLOT
381
0
42285
TrEMBL
-
A0A483BKJ8_OENOE
379
0
41661
TrEMBL
-
A0A1E3L2F1_9BACL
378
0
41178
TrEMBL
-
A0A653QI44_BACMY
377
0
41271
TrEMBL
-
J4WGM0_9ENTR
381
0
41611
TrEMBL
-
A0A653MQ66_9MICO
381
0
39742
TrEMBL
-
A0A7Y9PJZ6_9BACT
386
0
41508
TrEMBL
-
A0A7W4XN14_9PSED
381
0
42445
TrEMBL
-
H0U814_BRELA
375
0
40586
TrEMBL
-
A0A2X3LAK3_9ACTN
385
0
39212
TrEMBL
-
A0A284VKG4_9EURY
390
0
42445
TrEMBL
-
A0A0E1L1A1_CLOBO
380
0
41942
TrEMBL
-
A0A2U3L730_9DELT
401
0
43497
TrEMBL
-
A0A7W3NC99_9BACI
377
0
41247
TrEMBL
-
D5DSY6_PRIM1
Priestia megaterium (strain ATCC 12872 / QMB1551)
377
0
41217
TrEMBL
-
A0A653PZW0_9BACL
385
0
41436
TrEMBL
-
A0A2C9EPP2_PSEPH
Pseudomonas protegens (strain DSM 19095 / LMG 27888 / CFBP 6595 / CHA0)
424
0
45788
TrEMBL
-
A0A846MG45_9BACI
378
0
41286
TrEMBL
-
S6FTM3_9BACI
380
0
40729
TrEMBL
-
A0A653TAM9_9FLAO
402
0
43782
TrEMBL
-
A0A1S8TXF5_9CLOT
385
0
42559
TrEMBL
-
A0A7Y9NNQ5_9BACT
385
0
41203
TrEMBL
-
A0A852VFS9_9BACT
386
0
41332
TrEMBL
-
A0A0H3JB92_CLOPA
380
0
41810
TrEMBL
-
A0A653QHK7_MICLU
419
0
43639
TrEMBL
-
A0A653IBG7_9BACL
388
0
41817
TrEMBL
-
html completed


 results (
results ( results (
results ( top
top





