4.2.1.119: enoyl-CoA hydratase 2
This is an abbreviated version!
For detailed information about enoyl-CoA hydratase 2, go to the full flat file.
Reaction
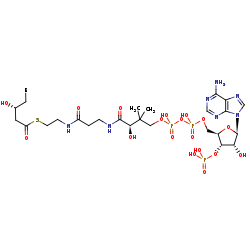 (3R)-3-hydroxyacyl-CoA=
(3R)-3-hydroxyacyl-CoA=
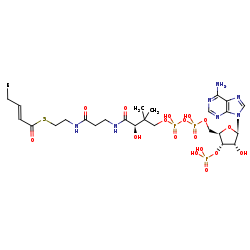 (2E)-2-enoyl-CoA +
(2E)-2-enoyl-CoA +
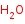 H2O
H2O
Synonyms
(3R)-hydroxyacyl-CoA dehydrogenase/2-enoyl-CoA hydratase 2, (R)-specific ECH, (R)-specific enoyl coenzyme A hydratase, (R)-specific enoyl-CoA hydratase, (R)-specific enoyl-coenzyme A hydratase, 2-enoyl-CoA hydratase, 2-enoyl-CoA hydratase 2, 2-enoyl-hydratase 2, 2E-enoyl-CoA hydratase 2, At1g76150, AtECH2, D-(-)-3-hydroxyacyl-CoA hydro-lyase, D-3-hydroxyacyl-CoA dehydratase, D-3-hydroxyacyl-CoA hydro-lyase, D-bifunctional enzyme, D-BP, D-specific 2-trans-enoyl-CoA hydratase, ECH-2, ECH2, enoyl-CoA hydratase 2, HFX_1483, hydratase 2, MaoC, MaoC-like protein, MFE-2, MFE2, MFE2 hydratase, Mfe2p [CtMfe2p(dha+bdelta)], multifunctional enzyme type 2, multifunctional enzyme type 2 hydratase, perMFE-II, peroxisomal enoyl-CoA hydratase 2, phaJ, PhaJ1, PhaJ1Pa, PhaJ1Pp, PhaJ4, PhaJ4aRe, PhaJ4bRe, PhaJ4cRe, PhaJAc, PhaJYB4, R-ECH, R-hydratase, R-specific enoyl coenzyme A hydratase, R-specific enoyl-CoA hydratase
ECTree
Specific Activity
Specific Activity on EC 4.2.1.119 - enoyl-CoA hydratase 2
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
0.05
-
L654G/V130G mutant with crotonyl-CoA as substrate, cell extract
0.08
-
L65A/V130G mutant with crotonyl-CoA as substrate, cell extract
0.12
-
strain HsMFE-2(D490A)
0.16
-
strain HsMFE-2(G16S)
0.2
-
strain HsMFE-2(H532A)
0.21
-
strain HsMFE-2(D370A), strain HsMFE-2(H406A)
0.24
-
strain HsMFE-2(Y410A)
0.26
-
strain HsMFE-2(D517A), strain HsMFE-2(E408A)
0.55
-
S62A mutant with octenoyl-CoA as substrate, cell extract
0.86
-
wild-type with octenoyl-CoA as substrate, cell extract
0.994
-
pH and temperature not specified in the publication
1.2
0.03 mM (2E)-hexadecenoyl-CoA as a substrate
1.98
-
L65G mutant with octenoyl-CoA as substrate, cell extract
12.4
0.1 mM, 3-hydroxydecanoyl-CoA as a substrate
1256
-
L65A mutant with crotonyl-CoA as substrate, cell extract
1288
-
V130A mutant with crotonyl-CoA as substrate, cell extract
15.8
-
L65G mutant with crotonyl-CoA as substrate, cell extract
1538
-
L65V mutant with crotonyl-CoA as substrate, cell extract
1594
-
wild-type with crotonyl-CoA as substrate, cell extract
1880
-
L65I mutant with crotonyl-CoA as substrate, cell extract
2.25
-
L65I mutant with octenoyl-CoA as substrate, cell extract
21.2
-
V130G mutant with octenoyl-CoA as substrate, cell extract
3.5
0.03 m (2E)-hexenoyl-CoA as a substrate
30
0.05 mM (2E)-decenoyl-CoA as a substrate
33.4
-
pET-Hydr2 expressed in Escherichia coli, soluble extract of the cells
48
-
recombinant 46 kDa hydratase 2, last purification step: size exclusion
6.59
-
V130A mutant with octenoyl-CoA as substrate, cell extract
67
-
S62A mutant with crotonyl-CoA as substrate, cell extract
68.5
-
V130G mutant with crotonyl-CoA as substrate, cell extract
69.8
-
L65A mutant with octenoyl-CoA as substrate, cell extract
7.92
-
L65V mutant with octenoyl-CoA as substrate, cell extract
883
-
V130G mutant with crotonyl-CoA as substrate, purified enzyme
additional information
-
activity is below the detection limit of the assay system when using extracts from non-transformed cells or cells transformed with the vector only




 results (
results ( results (
results ( top
top






