3.2.1.4: cellulase
This is an abbreviated version!
For detailed information about cellulase, go to the full flat file.
Reaction
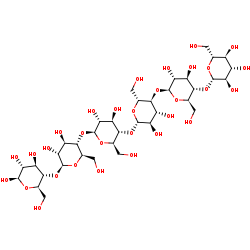 cellohexaose +
cellohexaose +
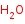 H2O=
2 cellotriose
H2O=
2 cellotriose
Synonyms
(1->4)-beta-D-glucan 4-glucanohydrolase, 1,4-beta-D-endoglucanase, 1,4-beta-D-glucan-4-glucanohydrolase, 168cel5, 9.5 cellulase, Abscission cellulase, AEG, Ag-EGase III, AgCMCase, alkali cellulase, Alkaline cellulase, AnCel5A, AtCel5, ATEG_07420, avicelase, BC-EG70a, Bc22Cel, BCE1, BcsZ, beta-1,4-endoglucan hydrolase, beta-1,4-endoglucanase, beta-1,4-glucanase, Bgl7A, bifunctional endoglucanase/xylanase, BlCel9, BP_Cel9A, Bx-ENG-1, Bx-ENG-2, Bx-ENG-3, C4endoII, carbomethyl cellulase, Carboxymethyl cellulase, Carboxymethyl-cellulase, carboxymethylcellulase, Cat 1, Caylase, CBH45-1, cbh6A, CBHI, CBHII, CcCel6C, CEL1, Cel1 EGase, Cel12A, Cel1753, Cel28a, Cel44A, Cel45A, Cel48A, Cel5, Cel5A, cel5B, Cel5E, Cel6A, Cel6A (E2), Cel6B, Cel6C, CEL7, Cel7A, Cel7B, Cel8, Cel8A, Cel8M, Cel8Y, Cel9A, CEL9A-50, CEL9A-65, Cel9A-68, CEL9A-82, Cel9A-90, Cel9B, CEL9C1, Cel9K, Cel9M, Cel9Q, CelA, CelB, CelC2 cellulase, CelCM3, CelDR, CelE, CelF, Celf_1230, Celf_3184, CelG, CelG endoglucanase, CelI15, cell-bound bacterial cellulase, CelL15, CelL73, cellic Ctec2, cellobiohydrolase, cellobiohydrolase I, celluase A, Celluclast, celludextrinase, Cellulase, cellulase 12A, cellulase A, cellulase A 3, cellulase Cel48F, cellulase Cel9A, cellulase Cel9M, cellulase CelC2, cellulase CelE, cellulase CM3, Cellulase E1, Cellulase E2, Cellulase E4, Cellulase E5, cellulase EGX, cellulase II, cellulase III, cellulase K, Cellulase SS, cellulase T, Cellulase V1, cellulase Xf818, cellulases I, cellulases III, cellulosin AP, Cellulysin, CelP, celS, CelStrep, celVA, CelX, CenA, CenC, CfCel6A, CfCel6C, CfEG3a, CHU_1280, CHU_2103, CjCel9A, Clocel_2741, CMCase, CMCase-I, CMCax, CMcellulase, Csac_1076, Csac_1078, CSCMCase, ctCel9D-Cel44A, CTendo45, ctendo7, CtGH5, Cthe_0435, CTHT_0045780, CX-cellulase, CyPB, DCC85_10145, DK-85, Dockerin type 1, Dtur_0671, E1 endoglucanase, Econase, EfPh, EG I, EG III, EG1, EG12, EG2, EG25, EG271, EG28, EG3, EG35, EG44, EG47, EG51, Eg5a, EG60, EGA, EGase, EGase II, EGB, EGC, EGCCA, EGCCC, EGCCD, EGCCF, EGCCG, EGD, EGE, EGF, egGH45, EGH, EGI, EGII, EGII/Cel5A, EGIV, EGL, EGL 1, Egl-257, Egl1, Egl499, Egl5a, EglA, eglB, EglC, EGLII, EglS, EGM, EGPf, EGPh, EGSS, EgV, EGX, EGY, EGZ, endo-1,4-B-glucanase, endo-1,4-beta-D-glucanase, endo-1,4-beta-glucanase, endo-1,4-beta-glucanase 1, endo-1,4-beta-glucanase 2, endo-1,4-beta-glucanase E1, endo-1,4-beta-glucanase V1, endo-beta-1,3-1,4-glucanase, endo-beta-1,4-glucanase, endo-beta-1,4-glucanase 1, endo-beta-1,4-glucanase 2, endo-beta-1,4-glucanase CMCax, endo-beta-1,4-glucanase EG27, endo-beta-1,4-glucanase EG45, endo-beta-D-1,4-glucanohydrolase, endo-beta-glucanase, endo-glucanase, ENDO1, ENDO2, endocellulase, endocellulase E1, endocellulases I, endocellulases II, endocellulases III, endocellulases IV, endogenous beta-1,4-endoglucanase, endogenous cellulase, endoglucanase, endoglucanase 1, endoglucanase 35, endoglucanase 47, endoglucanase CBP105, endoglucanase Cel 12A, endoglucanase Cel 5A, endoglucanase Cel 7B, endoglucanase Cel5A, endoglucanase Cel6A, endoglucanase D, endoglucanase EG-I, endoglucanase EG25, endoglucanase EG28, endoglucanase EG44, endoglucanase EG47, endoglucanase EG51, endoglucanase EG60, endoglucanase H, endoglucanase II, endoglucanase IIa, endoglucanase IV, endoglucanase L, endoglucanase M, endoglucanase V, endoglucanase Y, endolytic cellulase, EngA, EngH, EngL, EngM, engXCA, EngY, EngZ, family 7 cellobiohydrolase, FI-CMCASE, FnCel5A, FpCel45, Fpcel45a, GE40, GE40 endoglucanase, GH12 endo-1,4-beta-glucanase, GH124 endoglucanase, GH45 endoglucanase, gh45-1, GH5 cellulase, GH5 endoglucanase, GH6 endoglucanase, GH7 endoglucanase, GH9 termite cellulase, Glu1, Glu2, glycoside hydrolase family 9 endoglucanase, GtGH45, Lp-egl-1, manganese dependent endoglucanase, Maxazyme, Meicelase, mesophilic endoglucanase, mgCel6A, More, MtGH45, nmGH45, Onozuka R10, pancellase SS, PaPopCel1, PF0854, PH1171, PttCel9A, RCE1, RCE2, Roth 3056, RtGH124, Rucel5B, Sl-cel7, Sl-cel9C1, SnEG54, SoCel5, ssgluc, SSO1354, SSO1354 enzyme, SSO1354 protein, SSO1949, SSO2534, STCE1, Sumizyme, T12-GE40, TC Serva, TeEG-I, TeEgl5A, TfCel9A, ThEG, theme C glycoside hydrolase family 9 endo-beta-glucanase, Thermoactive cellulase, thermostable carboxymethyl cellulase, THITE_2110957, TM_1525, TM_1751, umcel5G, umcel9y-1, Vul_Cel5A
ECTree
Organism
Organism on EC 3.2.1.4 - cellulase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
-
-
-
brenda
-
-
-
brenda
CelJ
-
-
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
Actinomyces sp. 40 Korean Native Goat 40
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
aff. tenuissima, GenBank Accession: HQ630970
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
Uniprot
brenda
-
-
-
brenda
NO. F-50
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
i.e. Aspergillus fischeri
UniProt
brenda
i.e. Aspergillus fischeri
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
i.e. Eurotium herbariorum
UniProt
brenda
i.e. Eurotium herbariorum
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
IFO31125
-
-
brenda
-
-
-
brenda
cmc-1
-
-
brenda
isolated from rice bran
-
-
brenda
-
-
-
brenda
aff. fumigatus, GenBank Accession: HQ631021
-
-
brenda
-
-
-
brenda
GN1
-
-
brenda
-
UniProt
brenda
isolated from cow dung compost
-
-
brenda
isolated from cow dung compost
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
aff. pullans, GenBank Accession: HQ631013
-
-
brenda
-
-
-
brenda
Bacillus cellulyticus K-12
K-12
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
CTP-09
-
-
brenda
isolated from a cellulose-degrading enrichment culture initiated from compost samples
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
N-4
-
-
brenda
N-4 (NK1)
-
-
brenda
Bacillus sp. (in: Bacteria) No. 1139
No. 1139
-
-
brenda
isolated from Siderastrea stellata tissue samples collected at Cabo Branco shore, Brasil
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
BME-15
-
-
brenda
CK-2
-
-
brenda
DR
UniProt
brenda
isolated from the longtime-thermal compost containing rich cellulose materials in a factory of Zhengzhou in China, gene celI15
UniProt
brenda
JA18
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
isoform BC-EG70a
UniProt
brenda
Bellamya chinensis laeta UM-2014
isoform BC-EG70a
UniProt
brenda
-
-
-
brenda
GenBank Accession: HQ631009
-
-
brenda
acidophilic fungus, CGMCC 2500, gene bgl7A
UniProt
brenda
-
Uniprot
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
anaerobic fungal isolate, is purified from bovine rumen
-
-
brenda
-
-
-
brenda
-
-
-
brenda
strain NIAB 442 and its deoxyglucose- and streptomycin-resistant mutant 51 Smr
-
-
brenda
strain NIAB 442 and its deoxyglucose- and streptomycin-resistant mutant 51 Smr
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
CDBB 531 strain
SwissProt
brenda
CDBB 531 strain
SwissProt
brenda
-
-
-
brenda
-
-
-
brenda
aff. gramineum, GenBank Accession: HQ630973
-
-
brenda
-
-
-
brenda
-
-
-
brenda
GenBank Accession: HQ630978
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
aff. cladosporioides, GenBank Accession: HQ630971
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
Coleus scutellarioides
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
aff. bassiana, GenBank Accession: HQ630968
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
Ctenolepisma longicaudatum
hand-collected in buildings
-
-
brenda
isolated from a sample of a circumneutral geothermal pool at 94 °C
UniProt
brenda
bifunctional endo-1,4-beta-mannanase/endoglucanase, EC 3.2.1.78/EC 3.2.1.4, respectively
UniProt
brenda
bifunctional endo-1,4-beta-mannanase/endoglucanase, EC 3.2.1.78/EC 3.2.1.4, respectively
UniProt
brenda
-
-
-
brenda
GenBank Accession: HQ631008
-
-
brenda
aff. nigrum, GenBank Accession: HQ630972
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
S85
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
isolated from a cellulose-degrading enrichment culture initiated from compost samples
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
i.e. Polyporus tulipiferae
-
-
brenda
isolated from lime-applied garbage dumps
-
-
brenda
isolated from lime-applied garbage dumps
-
-
brenda
gene celF
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
Lenzites trabea
-
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
bifunctional CMCase and chitosanase, reactions of EC 3.2.1.4 and EC 3.2.16, respectively
UniProt
brenda
bifunctional CMCase and chitosanase, reactions of EC 3.2.1.4 and EC 3.2.16, respectively
UniProt
brenda
-
SwissProt
brenda
-
SwissProt
brenda
isolated from the marine sponge Dendrilla nigra
-
-
brenda
isolated from the marine sponge Dendrilla nigra
-
-
brenda
-
-
-
brenda
Japanese purple sea urchin
-
-
brenda
aff. bolleyi, GenBank Accession: HQ630981
-
-
brenda
GenBank Accession: HQ630974
-
-
brenda
NRRL 26519
-
-
brenda
NRRL 26519
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
F-2
-
-
brenda
KSM-N257
-
-
brenda
aff. oryzae, GenBank Accession: HQ630982
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
isolated from sediment
UniProt
brenda
-
UniProt
brenda
-
MG570051
GenBank
brenda
-
-
-
brenda
IBT 20888
-
-
brenda
IBT 20888
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
Phytomonas davidi
sandfly, Phlebotomus papatasi, infected
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
Cavara, pathogenic strain T-1 (Ken53-33) and nonpathogenic strain O(THU3*1), strain C-3(N87), strain N-1(H373)
-
-
brenda
-
UniProt
brenda
CBS814,70
-
-
brenda
-
UniProt
brenda
presence of two independent endogenous and symbiotic cellulolytic systems in flagellate-harboring termites
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
Y-20
-
-
brenda
Y-20
-
-
brenda
-
-
-
brenda
-
Uniprot
brenda
-
-
-
brenda
F-40
SwissProt
brenda
F40
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
member of glycoside hydrolase family 12
UniProt
brenda
putative; MT4 strain
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
JAM-KU023
-
-
brenda
JAM-KU023
-
-
brenda
-
UniProt
brenda
Mill.
-
-
brenda
aff. lignivora, GenBank Accession: HQ630984
-
-
brenda
Sporotrichum pulverulentum
Stachybotrys atra
-
-
-
brenda
-
-
-
brenda
NBRC 31817
UniProt
brenda
NBRC 31817
UniProt
brenda
-
UniProt
brenda
MTCC 1764
-
-
brenda
1326
-
-
brenda
1326
-
-
brenda
CMCase 1 and 2
-
-
brenda
A2
-
-
brenda
A2
-
-
brenda
contains two different types of CMCases type I and II
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
M23
-
-
brenda
BCC18080
Uniprot
brenda
BCC18080
Uniprot
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
i.e. Penicillium verruculosum
UniProt
brenda
i.e. Penicillium verruculosum
UniProt
brenda
i.e. Penicillium verruculosum
UniProt
brenda
i.e. Penicillium verruculosum
UniProt
brenda
i.e. Penicillium verruculosum
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
Cel6A
-
-
brenda
endoglucanase E-4 precursor; Cel9A
SwissProt
brenda
Thermochaetoides thermophila
Thermochaetoides thermophila CBS 144.50
-
UniProt
brenda
Thermochaetoides thermophila CT2
-
-
-
brenda
Thermochaetoides thermophila DSM 1495
-
UniProt
brenda
Thermochaetoides thermophila IMI 039719
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
DSM 27729
-
-
brenda
-
-
-
brenda
-
-
-
brenda
D-14
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
ATCC 48104
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
gene egl3
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
QM 9414, mutant strain
-
-
brenda
sandfly, Phlebotomus papatasi, infected
-
-
brenda
from a shallow marine hydrothermal vent from Vulcano Island
UniProt
brenda
-
-
-
brenda
from rumen of Bos mutus, Rucel5B
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
SwissProt
brenda
-
SwissProt
brenda
-
-
-
brenda
-
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
CelJ
-
-
brenda
deletion variants CelEM1, CelEM2 and CelEM3
-
-
brenda
endoglucanase 1b and 2
-
-
brenda
enzyme form EGA, EGB, EGC and EGD
-
-
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
production of several endoglucanases, which are part of a cellulase complex termed cellulosome
-
-
brenda
-
-
-
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
i.e. Ruminiclostridium thermocellum
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
multifunctional cellulase activity with endo-beta-1,4-glucanase activity, RC 3.2.1.4, exo-beta-1,4-glucanase activity, EC 3.2.9.11, and endo-beta-1,4-xylanase activity, EC 3.2.1.8
-
-
brenda
aff. phaeospermum, GenBank Accession: HQ630967
-
-
brenda
aff. sacchari, GenBank Accession: HQ630961
-
-
brenda
-
-
-
brenda
NO. F-50
-
-
brenda
-
-
-
brenda
Fresenius
-
-
brenda
isolates from hot spring water, sea water, soft wood, rice straw and cow dung
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
IFO31125
-
-
brenda
-
-
-
brenda
cmc-1
-
-
brenda
isolated from rice bran
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
GN1
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
CTP-09
-
-
brenda
isolated from a cellulose-degrading enrichment culture initiated from compost samples
-
-
brenda
N-4
-
-
brenda
N-4 (NK1)
-
-
brenda
No. 1139
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
BME-15
-
-
brenda
CelL15; isolated of feces sample of Golden Takin, Budorcas taxicolor, genes CelL15
UniProt
brenda
CelL73; isolated of feces sample of Golden Takin, Budorcas taxicolor, gene CelL73
UniProt
brenda
CK-2
-
-
brenda
DLG, N-24, or PAP115
-
-
brenda
DR
UniProt
brenda
isolated from the longtime-thermal compost containing rich cellulose materials in a factory of Zhengzhou in China, gene celI15
UniProt
brenda
JA18
-
-
brenda
CelL15; isolated of feces sample of Golden Takin, Budorcas taxicolor, genes CelL15
UniProt
brenda
CelL73; isolated of feces sample of Golden Takin, Budorcas taxicolor, gene CelL73
UniProt
brenda
isolated from a metagenomic library of a long-term dry thermophilic methanogenic digester community
UniProt
brenda
isolated from a thermophilic methanogenic digester community
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
bifunctional endo- and exoglucanase
UniProt
brenda
bifunctional enzyme, shows both endoglucanase and exoglucanase activities, EC 3.2.1.4 and 3.2.1.91, respectively
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
endoglucanase A and B
-
-
brenda
gene cenA
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
isoform Cel5B
UniProt
brenda
isoform Cel9C
UniProt
brenda
isoform Cel5B
UniProt
brenda
isoform Cel9C
UniProt
brenda
-
-
-
brenda
endoglucanase Z and Y
-
-
brenda
aff. salmonis, GenBank Accession: HQ630990
-
-
brenda
aff. spinifera, GenBank Accession: HQ631027
-
-
brenda
endoglucanase F
-
-
brenda
S85
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
isolated from a cellulose-degrading enrichment culture initiated from compost samples
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
ATCC 23769
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
ALKO4237
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
ALKO4237
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
F-2
-
-
brenda
KSM-N257
-
-
brenda
-
-
-
brenda
aff. minioluteum, GenBank Accession: HQ631007
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
endoglucanase family 5 protein
UniProt
brenda
HM003039; G5 CGMCC 3328
UniProt
brenda
HM003040; G5 CGMCC 3328
UniProt
brenda
aff. glomerata, GenBank Accession: HQ630999
-
-
brenda
aff. herbarum, GenBank Accession: HQ630963
-
-
brenda
-
-
-
brenda
var. cellulosa
-
-
brenda
wild-type and catabolite respression resistant mutants CRRmt4 and CRRmt24
-
-
brenda
-
SwissProt
brenda
-
UniProt
brenda
-
SwissProt
brenda
-
UniProt
brenda
-
SwissProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
CBS814,70
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
-
-
brenda
-
Uniprot
brenda
-
-
-
brenda
endoglucanase III
-
-
brenda
F-40
SwissProt
brenda
F40
-
-
brenda
-
UniProt
brenda
-
SwissProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
member of glycoside hydrolase family 12
UniProt
brenda
putative; MT4 strain
UniProt
brenda
strains P2, MT4, and Oalpha
UniProt
brenda
-
SwissProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
cv. Moneymaker
-
-
brenda
Mill.
-
-
brenda
cellulase I and II
-
-
brenda
QMB 482, ATCC 10011
-
-
brenda
Sporotrichum pulverulentum

-
-
-
brenda
Sporotrichum pulverulentum
ATCC 32629
-
-
brenda
Sporotrichum pulverulentum
five endoglucanases
-
-
brenda
-
-
-
brenda
M23
-
-
brenda
-
-
-
brenda
filamentous fungus, strain C-1, contains different cellulolytic enzymes
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
i.e. Penicillium verruculosum
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
-
-
brenda
a filamentous bacterium, strain ATCC BAA-629, genes Tfu_1627, Tfu_1074, Tfu_2176, and Tfu_0901, encoding endo-1,4-beta-glucanases
-
-
brenda
Cel6A
-
-
brenda
endoglucanase E-4 precursor; Cel9A
SwissProt
brenda
endoglucanase E1, E2, and E5
-
-
brenda
the bacterium produces six structurally and functionally distinct cellulases: Cel9B, Cel6A, Cel6B, Cel9A, Cel5A, Cel48A
-
-
brenda
Thermochaetoides thermophila

-
-
-
brenda
Thermochaetoides thermophila
-
UniProt
brenda
Thermochaetoides thermophila
-
UniProt
brenda
Thermochaetoides thermophila
var. coprophile
-
-
brenda
Thermochaetoides thermophila
var. dissitum
-
-
brenda
-
-
-
brenda
D-14
-
-
brenda
-
-
-
brenda
ATCC 48104
-
-
brenda
strain isolated from compost sample collected from Rohtak, Haryana, India
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
Swissprot
brenda
-
UniProt
brenda
-
Swissprot
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
Swissprot
brenda
-
UniProt
brenda
isoform Glu1
UniProt
brenda
isoform Glu2
UniProt
brenda
isoform Glu1
UniProt
brenda
isoform Glu2
UniProt
brenda
-
-
-
brenda
most efficient producer of cellulase
-
-
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
SwissProt
brenda
-
682688, 682986, 690562, 693480, 693939, 707205, 707226, 707818, 709689, 717549, 729011, 729404, 729411, 729655, 735430, 735466 -
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
ATCC 26921
-
-
brenda
gene egl3
-
-
brenda
i.e. Trichoderma reesei
-
-
brenda
QM6a
-
-
brenda
strains QM6a and PC-3-7, genes egl1 and egl3
-
-
brenda
-
UniProt
brenda
-
UniProt
brenda
QM6a
-
-
brenda
-
-
-
brenda
aff. atroviride, GenBank Accession: HQ630969
-
-
brenda
aff. koningii, GenBank Accession: HQ630959
-
-
brenda
-
-
-
brenda
a low-molecular weight enzyme form and a high-molecular weight enzyme form
-
-
brenda
cellulase II-A and II-B
-
-
brenda
QM 9414, mutant strain
-
-
brenda
-
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda
bacterial consortium SV79, isolated from sediments from Great Basin hot springs, Nevada, USA
-
-
brenda
isolated from a compost metagenome
-
-
brenda
isolated from camel rumen
UniProt
brenda
isolated from paddy soil
UniProt
brenda
isolated from saline-alkaline lake soil
UniProt
brenda
isolates of hyperthermophilic archaea from deep sea vents
UniProt
brenda
in rabbit caecum
-
-
brenda
rumen microbial metagenome library of goat rumen microorganisms
UniProt
brenda
-
-
-
brenda
-
UniProt
brenda









































 results (
results ( results (
results ( top
top






