3.2.1.136: glucuronoarabinoxylan endo-1,4-beta-xylanase
This is an abbreviated version!
For detailed information about glucuronoarabinoxylan endo-1,4-beta-xylanase, go to the full flat file.
Reaction
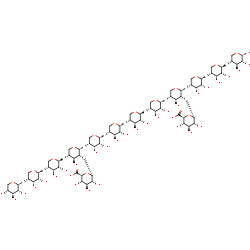 alpha-D-glucopyranuronosyl-(1-2)-[alpha-D-glucopyranuronosyl-(1-2)-[beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranose +
alpha-D-glucopyranuronosyl-(1-2)-[alpha-D-glucopyranuronosyl-(1-2)-[beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranose +
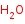 H2O=
H2O=
 4-O-beta-D-xylopyranosyl-beta-D-xylopyranose +
4-O-beta-D-xylopyranosyl-beta-D-xylopyranose +
 alpha-D-glucopyranuronosyl-(1-2)-[alpha-D-glucopyranuronosyl-(1-2)-[beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta--D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranose
alpha-D-glucopyranuronosyl-(1-2)-[alpha-D-glucopyranuronosyl-(1-2)-[beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta--D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranosyl-(1-4)]-beta-D-xylopyranosyl-(1-4)-beta-D-xylopyranose
Synonyms
1,4-beta-D-xylan xylanohydrolase, Agu115A, beta-1,4-endoxylanase, bifunctional cellulase/xylanase, endo-beta-xylanase, endoarabinoxylanase, endoglucanase, endoxylanase, feraxan endoxylanase, feraxanase, GH11 endoxylanase, GH115 alpha-glucuronidase, GH30 endoxylanase C, GH30 xylanase, glucuronoxylan xylanohydrolase, glucuronoxylan xylohydrolase, glucuronoxylan-specific xylanase A, glucuronoxylan-specific Xyn30D, glucuronoxylanase, glucuronoxylanase GH30, GuXN 30, M2, M4, TLX, Tx-Xyl, xylanase 30 A, xylanase C, xylanase, glucuronoarabinoxylan endo-1,4-beta-, Xyn30A, Xyn30D, XynA, XynC, XynGH30
ECTree
Sequence
Sequence on EC 3.2.1.136 - glucuronoarabinoxylan endo-1,4-beta-xylanase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
XYNC_BACIU
423
1
47840
Swiss-Prot
-
XYNC_BACSU
Bacillus subtilis (strain 168)
422
1
47337
Swiss-Prot
-
A0A7W7M937_9ACTN
556
1
58871
TrEMBL
-
A0A1J5SIK3_9ZZZZ
748
0
77411
TrEMBL
Secretory Pathway (Reliability: 4)
A0A6M4JGU4_BACSU
Bacillus subtilis (strain 168)
422
1
47337
TrEMBL
-
A0A1S8LJ70_9CLOT
582
0
63089
TrEMBL
-
W6SJL0_9CLOT
594
1
65973
TrEMBL
-
A0A1X9RHQ0_PAEAM
561
0
61622
TrEMBL
-
A0A1V5IYW4_9BACT
844
0
94080
TrEMBL
-
A0A0B0HRY5_9BACL
600
0
66313
TrEMBL
-
A0A1V5N1F0_9BACT
870
0
94862
TrEMBL
-
A0A110B050_9SPHI
491
0
52434
TrEMBL
-
A0A518BVL6_9BACT
Mucisphaera calidilacus
469
0
52464
TrEMBL
-
A0A5J4T4C6_9ZZZZ
486
0
54917
TrEMBL
Secretory Pathway (Reliability: 1)
A0A1V5NDG4_9BACT
452
0
50951
TrEMBL
-
A0A1V6J120_9BACT
469
0
51281
TrEMBL
-
A0A3N6C3R1_9ACTN
669
1
71082
TrEMBL
-
A0A1H7HB51_PHOVU
504
0
55605
TrEMBL
-
A0A840D6Y6_9BACE
545
0
60780
TrEMBL
-
A0A1J5SYZ3_9ZZZZ
483
0
54422
TrEMBL
Secretory Pathway (Reliability: 3)
A0A653RLR4_9BACI
420
1
47135
TrEMBL
-
G4HGL4_9BACL
422
1
47203
TrEMBL
-
A0A8F8Y4J5_9ACTN
581
0
63095
TrEMBL
-
A0A174KYP0_PHOVU
504
0
55593
TrEMBL
-
A0A1V5GHG4_9FIRM
860
0
93808
TrEMBL
-
A0A0P0FLE3_9BACE
546
0
61034
TrEMBL
-
A0A345H289_9FLAO
494
0
55923
TrEMBL
-
U5MRB7_CLOSA
423
0
47277
TrEMBL
-
E6UFE8_RUMA7
Ruminococcus albus (strain ATCC 27210 / DSM 20455 / JCM 14654 / NCDO 2250 / 7)
697
0
77097
TrEMBL
-
A0A653QJE5_9FLAO
489
0
52677
TrEMBL
-
A0A1A7KHA2_9FLAO
476
0
53027
TrEMBL
-
A0A1S8TAL1_9CLOT
423
1
47324
TrEMBL
-
A0A1V6EWX3_9BACT
470
0
53393
TrEMBL
-
A0A2M9N0A2_9BACL
426
1
47837
TrEMBL
-
A0A385ZPY3_9ACTN
490
0
52942
TrEMBL
-
M1MLA3_9CLOT
422
1
47323
TrEMBL
-
A0A199XPU4_9FLAO
489
0
52837
TrEMBL
-
A0A6C0QDS6_9ACTN
633
1
67602
TrEMBL
-
A0A1M7YU76_9VIBR
406
1
45322
TrEMBL
-
A0A5B9QFV5_9BACT
Bythopirellula goksoeyrii
727
0
78727
TrEMBL
-
A0A5P3AQ64_PHOVU
532
0
59492
TrEMBL
-
A0A6N2UZI1_9BACE
540
0
61055
TrEMBL
-
A0A1C5SCM1_9CLOT
uncultured Clostridium sp
448
0
51154
TrEMBL
-
A0A1M5XW15_9VIBR
406
0
45296
TrEMBL
-
A0A1N6M1X1_9VIBR
406
1
45422
TrEMBL
-
A0A4P8ETL7_BACSP
421
1
47279
TrEMBL
-
D2BZV8_DICZ5
Dickeya zeae (strain Ech586)
413
0
45227
TrEMBL
-
A0A1V5JMX2_9BACT
472
0
51579
TrEMBL
-
A0A841EZI5_9BACL
563
0
62066
TrEMBL
-
A0A650MIQ0_9CLOT
441
0
51420
TrEMBL
-
A0A381FCS3_9FLAO
476
0
53167
TrEMBL
-
A0A5P3AN37_PHOVU
504
0
55593
TrEMBL
-
A0A376E2Z6_9FLAO
477
0
53092
TrEMBL
-
A0A1Y6IYE3_9VIBR
466
0
52970
TrEMBL
-
A0A6F9ZFJ2_9BACT
504
0
55660
TrEMBL
-
A0A1V5T8X5_9SPIR
459
0
51447
TrEMBL
-
A0A0P0FFG4_BACT4
520
0
58566
TrEMBL
-
A0A139KK51_BACT4
520
0
59391
TrEMBL
-
A0A385ZXH1_9ACTN
446
0
48331
TrEMBL
-
M1N0D3_9CLOT
417
1
45438
TrEMBL
-
A0A199XNG4_9FLAO
486
0
54569
TrEMBL
-
A0A345H0B7_9FLAO
546
0
61343
TrEMBL
-
A0A0B0I0U8_9BACL
433
0
48062
TrEMBL
-
A0A0E4CYD3_9BACL
445
0
49009
TrEMBL
-
A0A654DGY8_SPHMU
506
0
54905
TrEMBL
-
C6C727_MUSP7
413
0
45592
TrEMBL
-
C6C727_MUSP7
Musicola paradisiaca (strain Ech703)
413
0
45592
TrEMBL
-
A0A2Z5JYJ1_9ACTN
470
0
50282
TrEMBL
-
A0A8F7SRN8_9BACI
422
1
47351
TrEMBL
-
A0A1S8LJE3_9CLOT
422
0
47121
TrEMBL
-
D3EH02_GEOS4
Geobacillus sp. (strain Y412MC10)
426
0
47831
TrEMBL
-
A0A7W4LS30_9BACI
422
1
47664
TrEMBL
-
E4T705_PALPW
Paludibacter propionicigenes (strain DSM 17365 / JCM 13257 / WB4)
423
0
46082
TrEMBL
-
A0A1R4LFJ9_9VIBR
406
1
45423
TrEMBL
-
A0A1V5L882_9ACTN
645
0
69956
TrEMBL
-
A0A5J4QB65_9ZZZZ
492
0
54639
TrEMBL
Secretory Pathway (Reliability: 1)
A0A1V5F7B1_9BACT
457
1
51269
TrEMBL
-
A0A1A8TI49_9GAMM
406
0
45587
TrEMBL
-
A0A0L6JL67_9FIRM
494
1
54710
TrEMBL
-
A0A1V5NCL4_9BACT
340
0
38172
TrEMBL
-
A0A5C0WM57_BACIA
428
1
48409
TrEMBL
-
A0A174SRZ0_BACT4
516
0
58258
TrEMBL
-
A0A2X2VNP1_9FLAO
474
0
52536
TrEMBL
-
A0A1V6CX68_9BACT
498
0
55522
TrEMBL
-
A0A3G4W6I5_9ACTN
668
1
71077
TrEMBL
-
A0A174MZ30_PHOVU
532
0
59510
TrEMBL
-
A0A1J5SG02_9ZZZZ
427
0
46149
TrEMBL
other Location (Reliability: 2)
A0A448U435_9FLAO
482
0
54722
TrEMBL
-
A0A1V5RV98_9BACT
474
0
54406
TrEMBL
-
A0A650MKB5_9CLOT
447
0
51853
TrEMBL
-
A0A1C6KMK6_9BACE
uncultured Bacteroides sp
540
0
61041
TrEMBL
-
A0A6N3D7Z4_9BACT
847
1
94057
TrEMBL
-
A0A498TUT0_BACIA
428
1
48453
TrEMBL
-
A0A1V6BHJ2_9BACT
486
1
53837
TrEMBL
-
G7M3Z8_9CLOT
416
1
44899
TrEMBL
-
A0A2M9MPP7_9BACL
561
0
61602
TrEMBL
-
A0A8F8U9N6_9ACTN
443
0
48521
TrEMBL
-
A0A448LGX4_9FLAO
474
0
53016
TrEMBL
-
M1MG04_9CLOT
423
1
46910
TrEMBL
-
A0A1V6F9C5_9BACT
447
0
51673
TrEMBL
-
A0A1V5LJH5_9BACT
433
0
48793
TrEMBL
-
A0A1V6J3X3_9BACT
475
0
52129
TrEMBL
-
A0A5P9HV30_9BACI
444
0
50422
TrEMBL
-
A0A1Q2Z1W9_9ACTN
422
0
46016
TrEMBL
-
A0A0L6JSW0_9FIRM
606
1
66392
TrEMBL
-
A0A853A2D3_9ACTN
648
1
68481
TrEMBL
-
A0A1V6J9L6_9BACT
488
0
55222
TrEMBL
-
A0A1C6EHZ9_9BACE
uncultured Bacteroides sp
546
0
61018
TrEMBL
-
A0A1V6BKG8_9BACT
443
0
50789
TrEMBL
-
A0A8I1WF89_BACIU
422
1
47403
TrEMBL
-
A0A100J065_9ACTN
422
0
46043
TrEMBL
-
A0A2X2KUV7_SPHMU
506
0
54951
TrEMBL
-
A0A1V5LFM5_9BACT
458
0
49947
TrEMBL
-
A0A1V6J5V1_9BACT
489
0
54813
TrEMBL
-
A0A2N7QYG9_9GAMM
536
1
58961
TrEMBL
-
A0A174W2Y9_PHOVU
336
0
36638
TrEMBL
-
A0A1S8M2P1_9CLOT
580
0
63094
TrEMBL
-
A0A653QKM9_9FLAO
486
0
54629
TrEMBL
-
M7N3A2_9BACT
490
0
53234
TrEMBL
-
A0A117EBU2_STRSC
434
1
47228
TrEMBL
-
A0A1C5WDN3_9BACE
uncultured Bacteroides sp
527
0
59870
TrEMBL
-
A0A174G6S0_PHOVU
504
0
55586
TrEMBL
-
A0A1S8TLU0_9CLOT
416
1
44944
TrEMBL
-
A0A1V6CTT1_9BACT
486
0
54955
TrEMBL
-
A0A5P9PLX6_9PSEU
462
0
48872
TrEMBL
-
A0A448AWW4_9FLAO
474
0
52563
TrEMBL
-
A0A090ZY18_PAEMA
447
0
49790
TrEMBL
-
F0S4T1_PSESL
Pseudopedobacter saltans (strain ATCC 51119 / DSM 12145 / JCM 21818 / CCUG 39354 / LMG 10337 / NBRC 100064 / NCIMB 13643)
510
0
56511
TrEMBL
-
S6FXS9_9BACI
422
1
47682
TrEMBL
-
A0A2K9EII0_9FIRM
533
1
59917
TrEMBL
-
A0A0P0GHA1_9BACE
540
0
61053
TrEMBL
-
A0A1V5FLM0_9BACT
421
0
46940
TrEMBL
-
A0A345H288_9FLAO
480
0
52745
TrEMBL
-
A0A174F697_9BACE
527
0
59890
TrEMBL
-
A0A377KZ28_ENTCA
495
0
56790
TrEMBL
-
A0A7W5DTV1_9PORP
405
0
44760
TrEMBL
-
A0A1Y6ITE6_9VIBR
406
1
45419
TrEMBL
-
A0A6H0H3F1_BACIU
422
1
47337
TrEMBL
-
A0A380CRQ8_SPHSI
498
0
54830
TrEMBL
-
A0A4U9JCE7_9SPHI
497
0
55166
TrEMBL
-
A0A846TXY1_9BURK
583
0
61772
TrEMBL
-
A0A1S8LHL1_9CLOT
518
0
57714
TrEMBL
-
A0A1S8LJI1_9CLOT
422
0
47035
TrEMBL
-
A0A174JRT8_9BACE
513
0
57838
TrEMBL
-
A0A1V6DW69_9BACT
469
1
52241
TrEMBL
-
A0A380ZKN1_PHOVU
504
0
55704
TrEMBL
-
A0A0P0GR67_9BACE
491
0
55217
TrEMBL
-
A0A1V2RBY5_9ACTN
437
0
47744
TrEMBL
-
A0A381FFG9_9FLAO
476
0
53129
TrEMBL
-
A0A1V5GUK9_9BACT
324
0
37173
TrEMBL
-
A0A1C6D4E0_9FIRM
uncultured Ruminococcus sp
952
1
106069
TrEMBL
-
A0A8I0PH53_9ACTN
434
1
46808
TrEMBL
-
A0A2P9F642_9ACTN
567
0
61289
TrEMBL
-
A0A5C5YU42_9BACT
630
1
69797
TrEMBL
-
A0A1W1HSX3_9BACT
492
0
55098
TrEMBL
-
A0A1V4SNL3_RUMHU
446
0
51113
TrEMBL
-
F2JKA9_CELLD
Cellulosilyticum lentocellum (strain ATCC 49066 / DSM 5427 / NCIMB 11756 / RHM5)
718
1
78900
TrEMBL
-
A0A5B9QRE5_9BACT
Bythopirellula goksoeyrii
681
1
75547
TrEMBL
-
A0A1V5ZE79_9BACT
353
0
40673
TrEMBL
-
A0A7X0M9Z4_9ACTN
542
0
57887
TrEMBL
-
A0A174N9G6_9BACE
492
0
55834
TrEMBL
-
A0A0U5F9F2_XANCI
109
0
11578
TrEMBL
-
A0A7W9LY66_9PSEU
422
0
46425
TrEMBL
-
A0A1V5GP27_9FIRM
771
0
83777
TrEMBL
-
A0A376J358_9FLAO
472
0
52416
TrEMBL
-
A0A4U8W8N1_9FLAO
487
0
52730
TrEMBL
-
M5RGV9_BACIT
420
1
47244
TrEMBL
-
A0A3S5E2P1_9FLAO
912
0
99672
TrEMBL
-
A0A1V4I9T0_9CLOT
444
0
50797
TrEMBL
-
A0A2N9LL56_9BACT
802
0
82614
TrEMBL
-
A0A7W9GIR0_9ACTN
425
1
46152
TrEMBL
-
A0A7Y9PJ42_9BACT
389
0
41652
TrEMBL
-
A0A1A7KHN5_9FLAO
479
0
54920
TrEMBL
-
A0A174Q997_BACUN
535
0
58663
TrEMBL
-
A0A1V6BZZ7_9BACT
494
0
54078
TrEMBL
-
A1XM14_9CELL
129
0
14542
TrEMBL
-
A3DJS9_ACET2
Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372)
630
0
70385
TrEMBL
Secretory Pathway (Reliability: 2)
H6WCZ0_PAEBA
562
0
61991
TrEMBL
-
Q46961_DICCH
413
0
45227
TrEMBL
-
F1TBY8_9FIRM
628
1
68198
TrEMBL
-
html completed



 results (
results ( results (
results ( top
top





