3.2.1.130: glycoprotein endo-alpha-1,2-mannosidase
This is an abbreviated version!
For detailed information about glycoprotein endo-alpha-1,2-mannosidase, go to the full flat file.
Word Map on EC 3.2.1.130 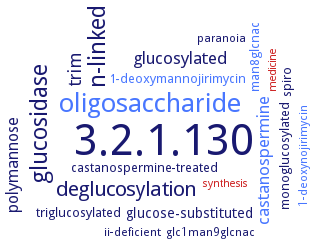
Reaction
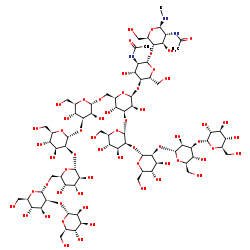 (Manalpha(1-2)Manalpha(1-6)(Manalpha(1-2)Manalpha(1-3))Manalpha(1-6))(Glcalpha(1-3)Manalpha(1-2)Manalpha(1-2)Manalpha(1-3))Manbeta(1-4)GlcNAcbeta(1-4)GlcNAc-R +
(Manalpha(1-2)Manalpha(1-6)(Manalpha(1-2)Manalpha(1-3))Manalpha(1-6))(Glcalpha(1-3)Manalpha(1-2)Manalpha(1-2)Manalpha(1-3))Manbeta(1-4)GlcNAcbeta(1-4)GlcNAc-R +
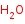 H2O=
H2O=
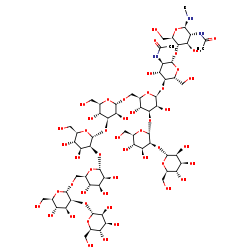 (Manalpha(1-2)Manalpha(1-6)(Manalpha(1-2)Manalpha(1-3))Manalpha(1-6))(Manalpha(1-2)Manalpha(1-3))Manbeta(1-4)GlcNAcbeta(1-4)GlcNAc-R +
(Manalpha(1-2)Manalpha(1-6)(Manalpha(1-2)Manalpha(1-3))Manalpha(1-6))(Manalpha(1-2)Manalpha(1-3))Manbeta(1-4)GlcNAcbeta(1-4)GlcNAc-R +
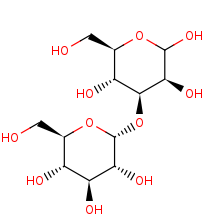 Glcalpha(1-3)Man
Glcalpha(1-3)Man
Synonyms
alpha-1,2-endomannosidase, alpha-endomannosidase, BtGH99, endo-alpha-1,2-mannanase, endo-alpha-1,2-mannosidase, endo-alpha-1->2-mannanase, endo-alpha-D-mannosidase, endo-alpha-mannosidase, endomannosidase, GH99, glucosyl mannosidase, glucosylmannosidase, Golgi endo-a-mannosidase, Golgi-endomannosidase, MANEA
ECTree
Sequence
Sequence on EC 3.2.1.130 - glycoprotein endo-alpha-1,2-mannosidase
Please wait a moment until all data is loaded. This message will disappear when all data is loaded.
Please wait a moment until the data is sorted. This message will disappear when the data is sorted.
MANEA_HUMAN
462
1
53671
Swiss-Prot
Secretory Pathway (Reliability: 2)
MANEA_MOUSE
462
1
53183
Swiss-Prot
Secretory Pathway (Reliability: 1)
MANEA_PONAB
462
1
53537
Swiss-Prot
Secretory Pathway (Reliability: 1)
MANEA_RAT
462
1
53416
Swiss-Prot
Secretory Pathway (Reliability: 1)
MANEA_XENLA
449
1
51862
Swiss-Prot
Secretory Pathway (Reliability: 2)
A0A841ND13_9FLAO
362
0
42211
TrEMBL
-
A0A6J8BDS9_MYTCO
620
1
73534
TrEMBL
Secretory Pathway (Reliability: 1)
A0A291SK03_STRMQ
371
0
40081
TrEMBL
-
A4IIA4_XENTR
432
1
49594
TrEMBL
Secretory Pathway (Reliability: 3)
A0A380KBR7_STREQ
724
0
82756
TrEMBL
-
A0A061IC84_CRIGR
491
1
56502
TrEMBL
other Location (Reliability: 4)
A0A380JY30_STRDY
228
1
25964
TrEMBL
-
M4YZB3_STREQ
724
0
82822
TrEMBL
-
K4QVU5_STRDJ
Streptomyces davaonensis (strain DSM 101723 / JCM 4913 / KCC S-0913 / 768)
713
0
75530
TrEMBL
-
A0A448DGN3_STREQ
724
0
82849
TrEMBL
-
F1ML11_BOVIN
465
1
54071
TrEMBL
Secretory Pathway (Reliability: 3)
I3VQI1_BOVIN
465
1
54087
TrEMBL
Secretory Pathway (Reliability: 2)
A0A7R8CFV7_LEPSM
399
0
46649
TrEMBL
Mitochondrion (Reliability: 5)
F9XHL5_ZYMTI
Zymoseptoria tritici (strain CBS 115943 / IPO323)
790
0
86918
TrEMBL
Secretory Pathway (Reliability: 1)
A0A6J8BA40_MYTCO
462
1
54027
TrEMBL
Secretory Pathway (Reliability: 1)
A0A8M1N255_DANRE
438
1
50607
TrEMBL
Secretory Pathway (Reliability: 1)
D2ATI5_STRRD
Streptosporangium roseum (strain ATCC 12428 / DSM 43021 / JCM 3005 / NI 9100)
510
1
53414
TrEMBL
-
A0A509DJK7_9STRE
724
0
82785
TrEMBL
-
G0PU30_9ACTN
723
1
76763
TrEMBL
-
C5WI64_STRDG
Streptococcus dysgalactiae subsp. equisimilis (strain GGS_124)
724
0
82846
TrEMBL
-
A0A829U7B7_STRCB
724
0
82340
TrEMBL
-
A0A0S2FC06_LYSAN
370
0
41795
TrEMBL
-
A0A8B6L7Z6_STREQ
724
0
82878
TrEMBL
-
A0A4V6LFZ1_STRDY
724
0
82848
TrEMBL
-
D6D1V7_9BACE
380
0
43437
TrEMBL
-
Q8A109_BACTN
Bacteroides thetaiotaomicron (strain ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50)
382
0
43215
TrEMBL
-
html completed




 results (
results ( results (
results ( top
top









